Compare samplers
In this notebook, we’ll compare the different samplers implemented in Bilby on a simple linear regression problem.
This is not an exhaustive set of the implemented samplers, nor of the settings available for each sampler.
Setup
[1]:
import bilby
import numpy as np
import matplotlib.pyplot as plt
from bilby.core.utils import random
# Sets seed of bilby's generator "rng" to "123" to ensure reproducibility
random.seed(123)
%matplotlib inline
[2]:
label = "linear_regression"
outdir = "outdir"
bilby.utils.check_directory_exists_and_if_not_mkdir(outdir)
Define our model
Here our model is a simple linear fit to some quantity \(y = m x + c\).
[3]:
def model(x, m, c):
return x * m + c
Simulate data
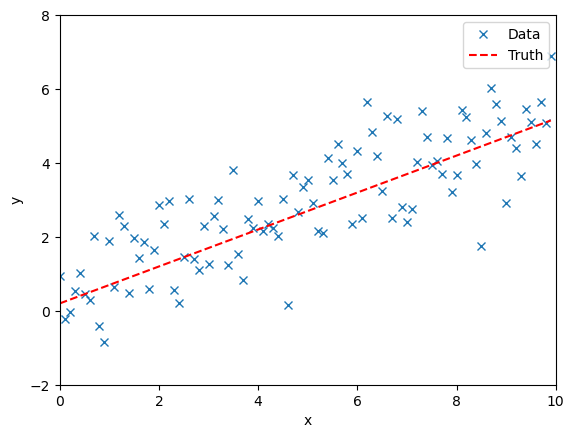
We simulate observational data. We assume some uncertainty in the observations and so perturb the observations from the truth.
[4]:
injection_parameters = dict(m=0.5, c=0.2)
sampling_frequency = 10
time_duration = 10
time = np.arange(0, time_duration, 1 / sampling_frequency)
N = len(time)
sigma = random.rng.normal(1, 0.01, N)
data = model(time, **injection_parameters) + random.rng.normal(0, sigma, N)
fig, ax = plt.subplots()
ax.plot(time, data, "x", label="Data")
ax.plot(time, model(time, **injection_parameters), "--r", label="Truth")
ax.set_xlim(0, 10)
ax.set_ylim(-2, 8)
ax.set_xlabel("x")
ax.set_ylabel("y")
ax.legend()
plt.show()
plt.close()
Define the likelihood and prior
For any Bayesian calculation we need a likelihood and a prior.
In this case, we take a GausianLikelihood as we assume the uncertainty on the data is normally distributed.
For both of our parameters we take uniform priors.
[5]:
likelihood = bilby.likelihood.GaussianLikelihood(time, data, model, sigma)
priors = bilby.core.prior.PriorDict()
priors["m"] = bilby.core.prior.Uniform(0, 5, "m")
priors["c"] = bilby.core.prior.Uniform(-2, 2, "c")
Run the samplers and compare the inferred posteriors
We’ll use four of the implemented samplers.
For each one we specify a set of parameters.
Bilby/the underlying samplers produce quite a lot of output while the samplers are running so we will suppress as many of these as possible.
After running the analysis, we print a final summary for each of the samplers.
[6]:
samplers = dict(
bilby_mcmc=dict(
nsamples=1000,
L1steps=20,
ntemps=10,
printdt=10,
),
dynesty=dict(npoints=500, sample="acceptance-walk", naccept=20),
pymultinest=dict(nlive=500),
nestle=dict(nlive=500),
emcee=dict(nwalkers=20, iterations=500),
ptemcee=dict(ntemps=10, nwalkers=20, nsamples=1000),
)
results = dict()
[7]:
bilby.core.utils.logger.setLevel("ERROR")
for sampler in samplers:
result = bilby.core.sampler.run_sampler(
likelihood,
priors=priors,
sampler=sampler,
label=sampler,
resume=False,
clean=True,
verbose=False,
**samplers[sampler]
)
results[sampler] = result
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/sampler/pymultinest.py:43: FutureWarning: The PyMultiNest sampler interface in bilby is deprecated and will be removed in Bilby version 3. Please use the `pymultinest-bilby`sampler plugin instead: https://github.com/bilby-dev/pymultinest-bilby.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
*****************************************************
MultiNest v3.10
Copyright Farhan Feroz & Mike Hobson
Release Jul 2015
no. of live points = 500
dimensionality = 2
*****************************************************
analysing data from /tmp/tmp0tyl36b8/.txt ln(ev)= -140.53693139345233 +/- 0.10723620363507863
Total Likelihood Evaluations: 5575
Sampling finished. Exiting MultiNest
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/sampler/nestle.py:34: FutureWarning: The Nestle sampler interface in bilby is deprecated and will be removed in Bilby version 3. Please use the `nestle-bilby`sampler plugin instead: https://github.com/bilby-dev/nestle-bilby.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/sampler/ptemcee.py:134: FutureWarning: The ptemcee sampler interface in bilby is deprecated and will be removed in Bilby version 3. Please use the `ptemcee-bilby`sampler plugin instead: https://github.com/bilby-dev/ptemcee-bilby.
warnings.warn(msg, FutureWarning)
/opt/conda/envs/python311/lib/python3.11/site-packages/bilby/core/likelihood.py:113: FutureWarning: Parameter attribute queried for <class 'bilby.core.likelihood.GaussianLikelihood'>. This is deprecated behaviour and will be removed in Bilby version 3. See https://bilby-dev.github.io/bilby/parameters for more details.
warnings.warn(msg, FutureWarning)
[8]:
print("=" * 40)
for sampler in results:
print(sampler)
print("=" * 40)
print(results[sampler])
print("=" * 40)
========================================
bilby_mcmc
========================================
nsamples: 2141
ln_noise_evidence: nan
ln_evidence: -137.678 +/- 0.029
ln_bayes_factor: nan +/- 0.029
========================================
dynesty
========================================
nsamples: 1287
ln_noise_evidence: nan
ln_evidence: -140.451 +/- 0.135
ln_bayes_factor: nan +/- 0.135
========================================
pymultinest
========================================
nsamples: 1781
ln_noise_evidence: nan
ln_evidence: -140.537 +/- 0.107
ln_bayes_factor: nan +/- 0.107
========================================
nestle
========================================
nsamples: 4274
ln_noise_evidence: nan
ln_evidence: -140.411 +/- 0.107
ln_bayes_factor: nan +/- 0.107
========================================
emcee
========================================
nsamples: 8560
ln_noise_evidence: nan
ln_evidence: nan +/- nan
ln_bayes_factor: nan +/- nan
========================================
ptemcee
========================================
nsamples: 1020
ln_noise_evidence: nan
ln_evidence: -144.445 +/- 20.167
ln_bayes_factor: nan +/- 20.167
========================================
Make comparison plots
We will make two standard comparison plots.
In the first we plot the one- and two-dimensional marginal posterior distributions in a “corner” plot.
In the second, we show the inferred model that we are fitting along with the uncertainty by taking random draws from the posterior distribution. This kind of posterior predicitive plot is useful to identify model misspecification.
[9]:
_ = bilby.core.result.plot_multiple(
list(results.values()), labels=list(results.keys()), save=False
)
plt.show()
plt.close()
/opt/conda/envs/python311/lib/python3.11/site-packages/arviz/__init__.py:50: FutureWarning:
ArviZ is undergoing a major refactor to improve flexibility and extensibility while maintaining a user-friendly interface.
Some upcoming changes may be backward incompatible.
For details and migration guidance, visit: https://python.arviz.org/en/latest/user_guide/migration_guide.html
warn(
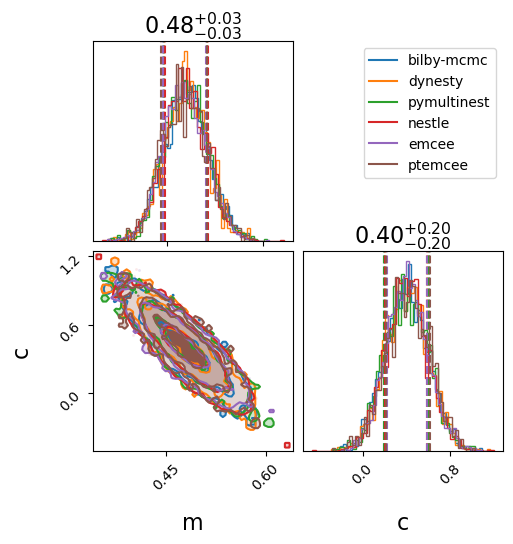
[10]:
fig, ax = plt.subplots(figsize=(12, 8))
ax.plot(time, data, "x", label="Data", color="r")
ax.plot(
time, model(time, **injection_parameters), linestyle="--", color="k", label="Truth"
)
for jj, sampler in enumerate(samplers):
result = results[sampler]
samples = result.posterior[result.search_parameter_keys].sample(500)
for ii in range(len(samples)):
parameters = dict(samples.iloc[ii])
plt.plot(time, model(time, **parameters), color=f"C{jj}", alpha=0.01)
plt.axhline(-10, color=f"C{jj}", label=sampler.replace("_", " "))
ax.set_xlim(0, 10)
ax.set_ylim(-2, 8)
ax.set_xlabel("Time")
ax.set_ylabel("y")
ax.legend(loc="upper left")
plt.show()
plt.close()
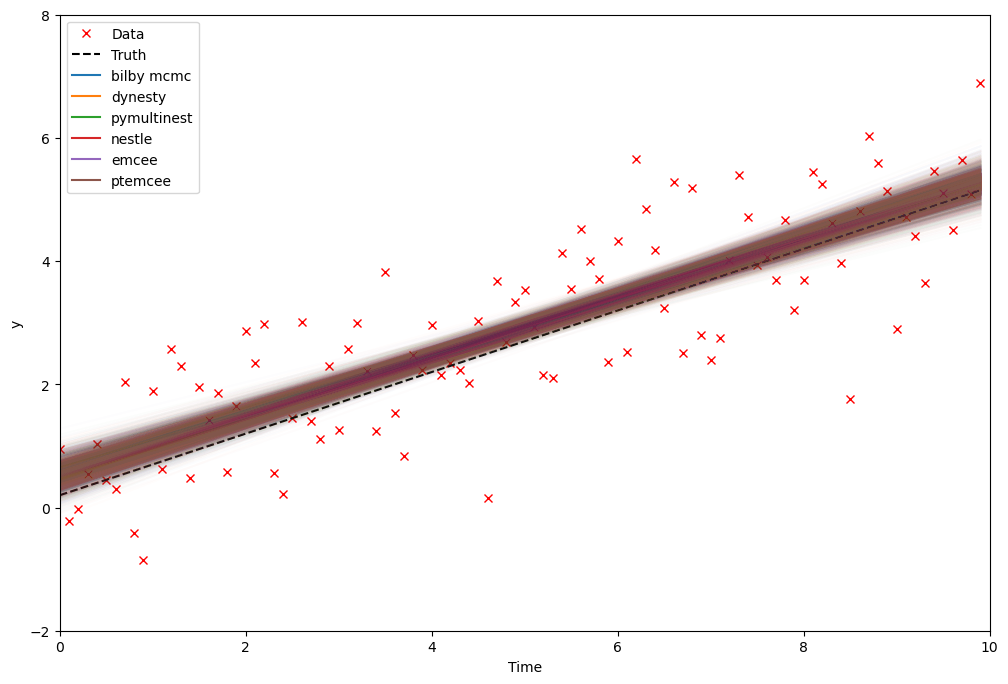
[ ]: